Leave message
Can’t find what you’re looking for?
Fill out this form to inquire about our custom protein services!
Inquire about our Custom Services >>































 Request a FREE Sample of our FcRn Binding Kit!
Request a FREE Sample of our FcRn Binding Kit! Request a FREE Sample of our Fc gamma RI / CD64 Binding Kit !
Request a FREE Sample of our Fc gamma RI / CD64 Binding Kit !
 Happy Holiday! Limited Keychain here with your next order
Happy Holiday! Limited Keychain here with your next order Happy Holiday! Limited Keychain here with your next order
Happy Holiday! Limited Keychain here with your next order
 Request a FREE sample of our GMP products!
Request a FREE sample of our GMP products!  Request a FREE sample of our GMP products!
Request a FREE sample of our GMP products!
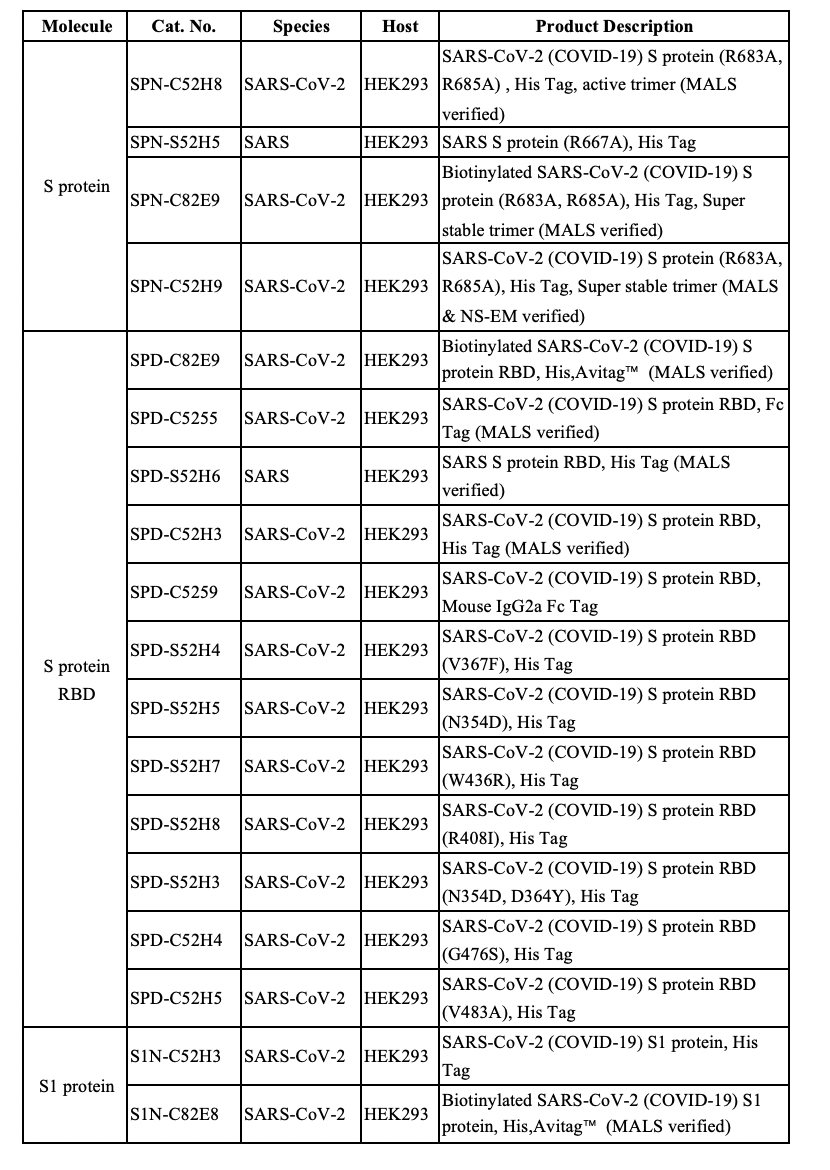
The trimeric spike protein (S) plays a key role in the early steps of COVID-19 infection. The S1 domain is responsible for receptor binding and the S2 domain mediating membrane fusion.[1-3] Triggered by the receptor binding, the spike protein undergoes a transition from a metastable prefusion state to a stable post-fusion state during infection, in which the receptor-binding subunit is cleaved by the protease like TMPRSS2, and the fusion subunit undergoes large-scale conformational rearrangements to expose the hydrophobic fusion peptide.[4,5] Thus, the receptor-binding domain (RBD) has become a hot target for therapeutic development against COVID-19.
There is another part of the S1 protein called N terminal domain (NTD). Although the function of NTD is not well characterized, in some coronaviruses, the NTD may recognize specific sugar moieties upon initial attachment and play a role during the infection.[6-8] Thus, RBD should not be treated as the only target, and the NTD could potentially become a target for SARS-CoV-2 as well.[9]
Recently Dr. Chen’s team from China published their work in Science and supported this idea. They isolated human mAbs from 10 Chinese patients recovered from COVID-19 infection. In their studies, there was a mAb with a high affinity to SARS-CoV-2 RBD but failed to neutralize authentic SARS-CoV-2. There is also another NTD binding mAb (4A8) exhibiting a high level of neutralization against SARS-CoV-2 in vitro but does not block the interaction between ACE2 and S protein (Figure 1). Their work suggests the presence of other important mechanisms for SARS-CoV-2 neutralization in addition to suppressing the viral interaction with the receptor. Scientists should consider the whole Spike protein instead of just RBD when developing therapeutics. Combination of the NTD targeting antibodies and RBD targeting antibodies as the promising cocktail therapy may avoid the escaping mutations of virus.[10]
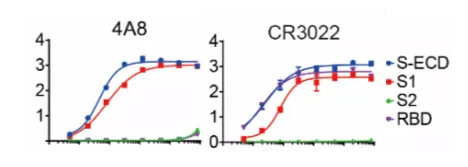
Figure 1. Binding curves of representative mAbs.
ACROBiosystems developed SARS-CoV-2 S1 NTD protein (Cat. No. S1D-C52H6), which can specifically bind to anti-SARS-CoV-2 S1 NTD antibody (Cat. No. SPD-M121). The bioactivity of the S1 NTD protein has been verified through ELISA, BLI and SPR assays. They are perfect for immunization, screening and characterization of therapeutic development.
Assay Data
>>> ELISA
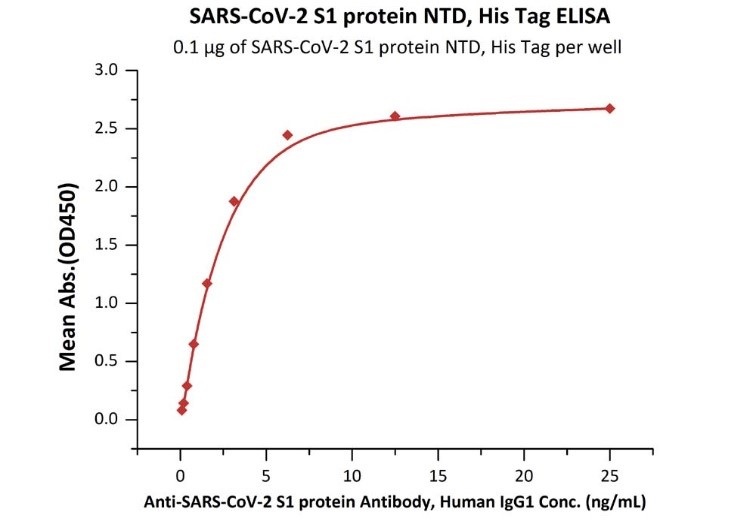
Figure.1 Immobilized SARS-CoV-2 S1 protein NTD, His Tag (Cat. No. S1D-C52H6) at 1 μg/mL (100 μL/well) can bind Anti-SARS-CoV-2 Spike NTD Antibody, Chimeric mAb (Cat. No. SPD-M121) with a linear range of 0.1-3 ng/mL.
>>> BLI
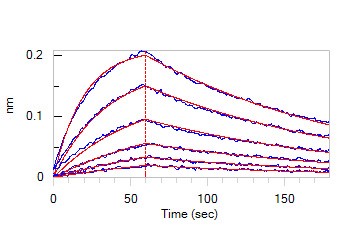
Figure. 2 Loaded Anti-SARS-CoV-2 Spike NTD Antibody, Chimeric mAb (Cat. No. SPD-M121) on AHC Biosensor, can bind SARS-CoV-2 S1 protein NTD, His Tag (Cat. No. S1D-C52H6) with an affinity constant of 39.8 nM as determined in BLI assay (ForteBio Octet Red96e)
>>> SPR
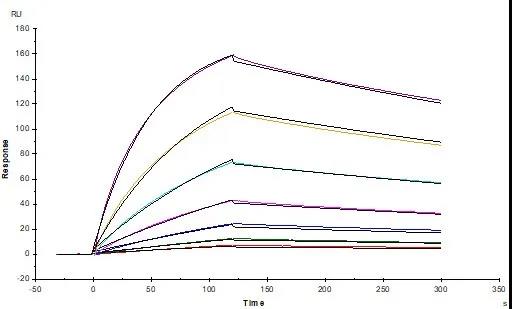
Figure. 3 Anti-SARS-CoV-2 Spike NTD Antibody, Chimeric mAb (Cat. No. SPD-M121) captured on CM5 chip via Anti-human IgG Fc antibodies surface can bind SARS-CoV-2 S1 protein NTD, His Tag (Cat. No. S1D-C52H6) with an affinity constant of 74.6 nM as determined in a SPR assay (Biacore T200)
All Spike Protein List
Reference
1. Gallagher TM, Buchmeier MJ, Coronavirus spike proteins in viral entry and pathogenesis, Virology, 2001 Jan 20; 279(2):371-4.
2. Simmons G, et al., Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research, Antiviral Res. 2013 Dec; 100(3):605-14.
3. Belouzard S, et al., Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites, Proc Natl Acad Sci USA 2009 Apr 7; 106(14):5871-6.
4. Song W, et al., Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2, PLoS Pathog. 2018 Aug; 14(8): e1007236.
5. Hoffmann M, et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, 2020 Apr 16; 181(2):271-280.e8.
6. Wrapp D, et al., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science, 2020 Mar 13; 367(6483):1260-1263.
7. Walls AC, et al., Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein, Cell, 2020 Apr 16; 181(2):281-292.e6.
8. Krempl C, et al., Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus, J Virol. 1997 Apr; 71(4):3285-7.
9. Zhou H, et al., Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein, Nat Commun. 2019 Jul 11; 10(1):3068.
10. Chi X. et al., A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020, DOI: 10.1126/science.abc6952
This web search service is supported by Google Inc.